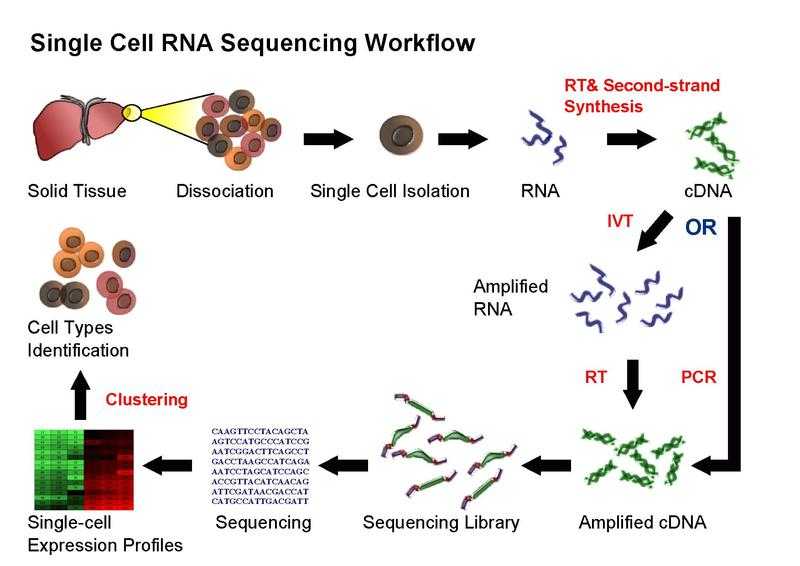
Transcriptional similarity has been used to differentiate and categorize cells within a tissue, it also gives insights about functions of a cell.
Similarly, we use single cell data to identify cell-type-specific markers and then label and map these cell types back in the intact tissue.
Conventional ‘bulk’ methods of RNA sequencing (RNA-seq) process hundreds of thousands of cells at a time and average out the differences. But no two cells are exactly alike, and scRNA-seq can reveal the subtle changes that make each one unique. It can even reveal entirely new cell types.
For example, one study identified many rare cell types in the gut that likely function in secretion (Nature 525, 251–255, 2015).
For instance, after using scRNA-seq to probe some 2,400 immune-system cells, Aviv Regev of the Broad Institute in Cambridge, Massachusetts, and her colleagues came across some dendritic cells that had potent T-cell-stimulating activity (A.-C. Villani et al. Science 356, eaah4573; 2017). Regev says that a vaccine to stimulate these cells could potentially boost the immune system and protect against cancer.
But why such discoveries are hard won?
- It is difficult to manipulate a single cell than large population.
- High accuracy needed as a single cell has a tiny amount of RNA.
- Analysis of large amount of data.
- Data and consensus sequences are unknown now.
But a range of online resources and tools are beginning to ease the process of scRNA-seq data analysis. One page at GitHub, called ‘Awesome Single Cell’ (go.nature.com/2rmb1hp), catalogues more than 70 tools and resources, covering every step of the analysis process.
Several clinical applications will emerge for scRNA-seq in the next 5 or so years. For example, resected tumours might be routinely assessed for the presence of rare malignant and chemo-resistant cancer cells. This information will provide crucial diagnostic information and will guide decisions regarding treatment. Next, as an extension to a full blood count, scRNA-seq assessments will provide in-depth information on the response of immune cells, which again will inform diagnoses and the choice of therapy. Finally, the relatively small numbers of cells present in a range of other tissue biopsies, for example from the skin and gut mucosal surfaces, will be ideal for providing molecular data that informs on diagnosis, disease progression and appropriate treatments. Thus, scRNA-seq will progress out of specialist research laboratories and will become an established tool for both basic scientists and clinicians alike.
Ref:
https://genomemedicine.biomedcentral.com/articles/10.1186/s13073-017-0467-4
https://www.nature.com/news/single-cell-sequencing-made-simple-1.22233